|
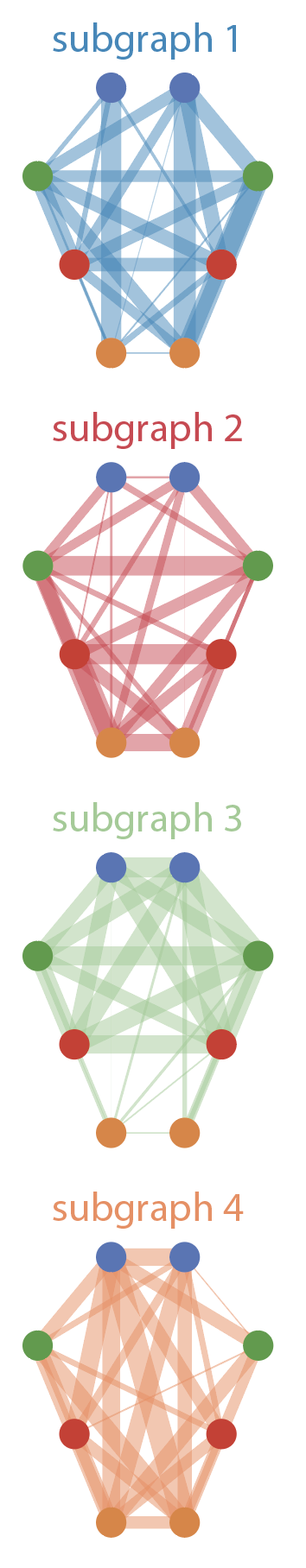
[NMF Analysis] Using a sliding window approach, we can identify time-varying functional connectivity, using simple correlation across the anatomical regions estimated within each time window. |
[Left] We can represent factors derived from NMF as subnetworks that are variably expressed over time. Networks derived from a four-factor factorisation are shown here. [Middle] The factors' subnetwork expression varies over time, but shows persistent differences across genotypes. [Right] Full time varying connectivity matrices are shown here, with time points corresponding to the dashed line in the middle plot. |